Research
Research
The intersection between photons and proteins is exciting! Biologically captured photons are the source of almost all chemical fuel on Earth -- in either biomass or fossil state. Photoactive proteins let us see, control neural circuits, and can repair DNA.
Our group aims to use the unique hardware and computational facilities of DESY to study the intersection of light and biology. In turn, the science questions we pose will help inform the next large-scale investments from the laboratory to the facility scale.
Measuring Correlated Motions in Proteins
We know proteins make use of long-range motions to perform the essential functions of life: turn light into chemical work, transmit signals, conduct chemistry, and mechanically alter the environment. How proteins accomplish these amazing tasks remains unexplained at the atomic level, largely because experimentally measuring these long-range motions at atomic detail has been a challenge.
To address this, we've investigated protein crystal diffuse scatter, caused by disorder in protein crystals. If, for example, proteins were trapped in a crystal in two (or more) different functional states this should appear as the diffuse scatter and be measurable! Check out our publication in IUCrJ and the associated commentary.
Recently, via a different technique, our group had success measuring structural correlations in the main protease (Mpro) from SARS-COV-2. By analyzing structural variations across a large number of protein crystals, we are able to see correlations between the active site of the protein and the dimer interface 30+ Å away. The enzyme is only functional as a dimer, and perhaps we are seeing the first glimpses of why... this is new and ongoing work we are excited to be pursuing it!
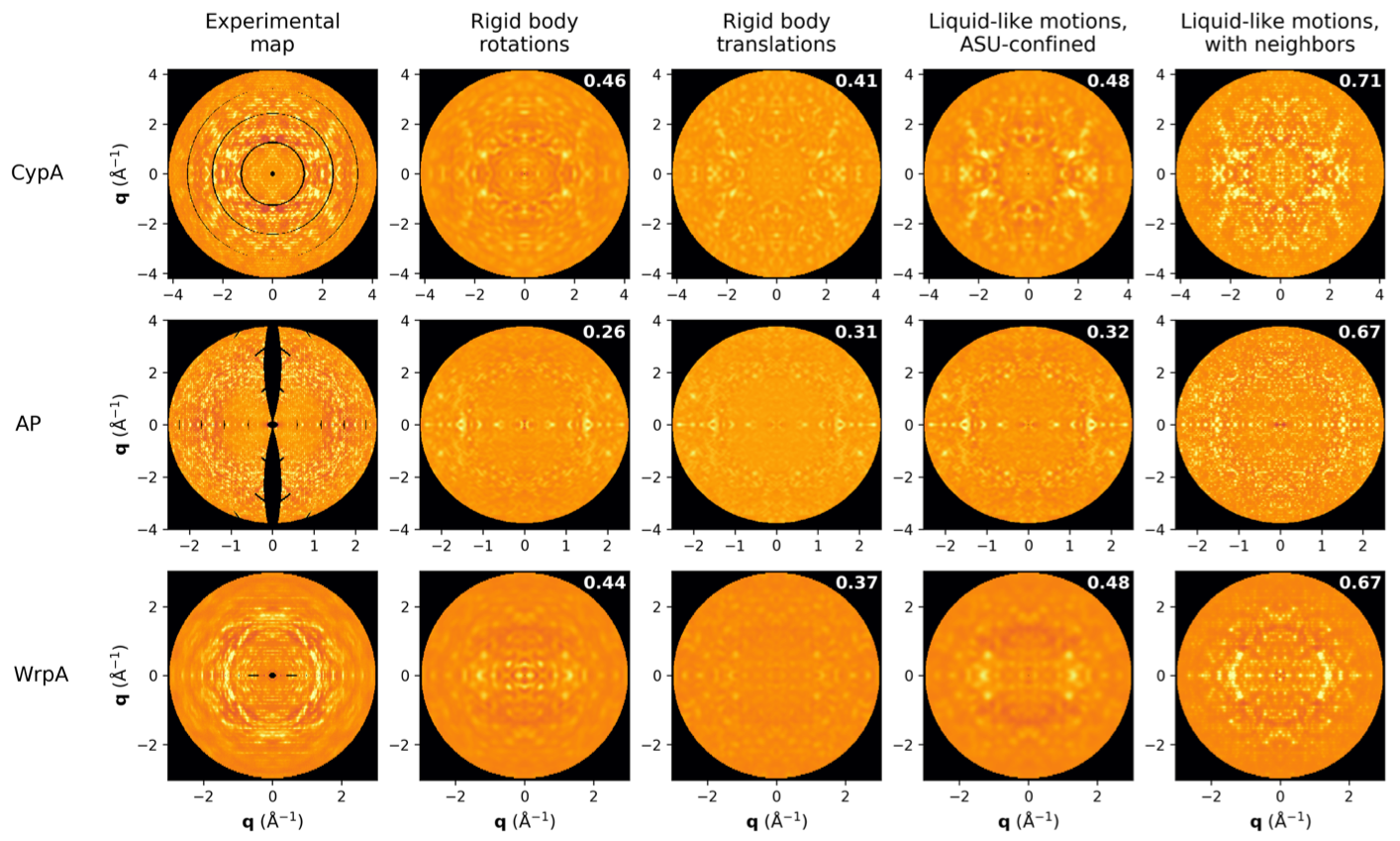
The diffuse scatter is a rich signal that reports on correlated disorder in crystals. Since functional motions such as ligand binding are such correlation motions, we hope to push the analysis of diffuse scatter to include the extraction of these biologically relevant signals. So far the best models -- shown here up against experimental data for 3 model proteins -- report on the basic physical properties of protein crystals. Adding correlations that extend beyond a single unit cell boundary dramatically increases the quality of the model, both quantitatively (correlation coefficients shown) and qualitatively.
Time-resolved Serial Crystallography of Photoenzymes
Photoenzyme systems represent a unique opportunity to tackle fundamental questions about the origins of enzyme catalysis. Catalysis in photoenzyme systems can be triggered with femtosecond lasers and the resulting dynamics imaged using the new technique of serial femtosecond crystallography (SFX). Enabled by the availability of hard x-ray free electron lasers (XFELs), this technique involves capturing x-ray images from a large number of protein crystals (10,000+) that are combined to form a 3D map of the ensemble average atomic positions. By changing the pump (laser)/probe (x-ray) delay, a time course can be obtained, tracking the atomic positions over time, with a resolution of about 30 fs in modern setups. SFX is less than a decade old but has already allowed the filming of photoactive non-enzyme proteins at Å/fs resolution. Extending the technique to photoenzyme systems – that do chemistry – will enable us to address specific questions about how enzymes do catalysis.
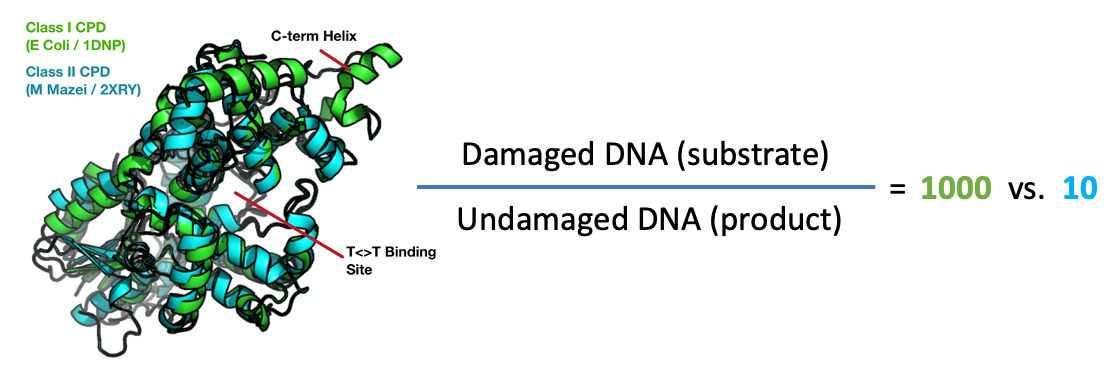
Photolyase repairs DNA damage using blue light and is a classic photoenzyme system. One interesting question is how photolyase discriminates between damaged DNA and normal (repaired) DNA. Indeed, the ability for the enzyme find damage sites and then release repaired DNA after it's job is complete is the rate-limiting step in the enzyme's function!
Shown here are the crystal structures of the photolyase from E. Coli (green) and M. Mazei (teal), which are examples of class I and class II photolyases, respectively. The two proteins share the same fold and conduct the same chemistry despite being only 24% identical (ialign algorithm). The class I CPD is much more discriminating, binding damaged DNA 1000x tighter than undamaged sequences, as compared to a 10x preference for the class II structure. It seems likely that this additional discriminatory power is due to the major structural difference between the two proteins, an additional c- terminal alpha helix found in class I structures (indicated). Perhaps the dynamics of these systems can tell us why this difference occures. We are pursuing time-resolved experiments to resolve this mystery and many others!
Photo-triggering Enzymes
One very new avenue of research in our group: can we chemically modify enzymes, their substrates, or their co-factors to make them photo-triggerable? Previous work in this area has focused on photo-caged species, where the natural compound is released or generated after photon absorption. We want to do something a bit different: create analogs of the natural species that are intrinsically photon-activated.
The key advantage of the latter strategy is that photoactive compounds can react very quickly, down to femtosecond timescales -- the natural timescales on which chemistry and catalysis (ie, speeding up chemistry) occur. Caged compounds rely on diffusion and other slow processes to work, precluding the study of the electronic rearrangements involved in bond making and breaking.
If you are interested in learning more, get in touch!

This is a new direction we are excited about that is supported by our collaborators and friends at HARBOR, a new building on the DESY campus dedicated to the study of ultrafast processes in biology. HARBOR offers access to laser systems, synthesis labs, and protein expression/purification/crystallization equipment all under one roof.